| Chlamydomonas reinhardtii | |
|---|---|
 | |
| Scientific classification | |
| Kingdom: | Plantae |
| Phylum: | Chlorophyta |
| Class: | Chlorophyceae |
| Order: | Chlamydomonadales |
| Family: | Chlamydomonadaceae |
| Genus: | Chlamydomonas |
| Species: | C. reinhardtii |
| Binomial name | |
| Chlamydomonas reinhardtii P.A.Dang. | |
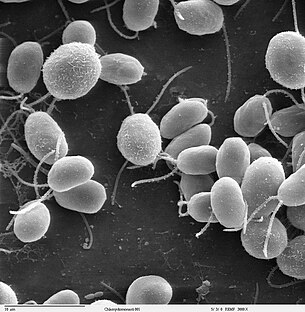
Chlamydomonas reinhardtii is a single-cell green alga about 10 micrometres in diameter that swims with twoflagella. It has a cell wall made of hydroxyproline-rich glycoproteins, a large cup-shaped chloroplast, a largepyrenoid, and an "eyespot" that senses light.
Chlamydomonas species are widely distributed worldwide in soil and fresh water. Chlamydomonas reinhardtii is an especially well studied biological model organism, partly due to its ease of culturing and the ability to manipulate its genetics. When illuminated, C. reinhardtii can grow photoautotrophically, but it can also grow in the dark if supplied with organic carbon. Commercially, C. reinhardtii is of interest for producing biopharmaceuticals and biofuel, as well being a valuable research tool in making hydrogen.
Contents
History
The C. reinhardtii wild-type laboratory strain c137 (mt+) originates from an isolate made near Amherst, Massachusetts, in 1945 by Gilbert M. Smith.
The species has been spelled several different ways because of different transliterations of the name from Russian: reinhardi, reinhardii, and reinhardtii all refer to the same species, C. reinhardtii Dangeard.
Model organism
Chlamydomonas is used as a model organism for research on fundamental questions in cell and molecular biology such as:
- How do cells move?
- How do cells respond to light?
- How do cells recognize one another?
- How do cells generate regular, repeatable flagellar waveforms?
- How do cells regulate their proteome to control flagellar length?
- How do cells respond to changes in mineral nutrition? (nitrogen, sulfur, etc.)
There are many known mutants of C. reinhardtii. These mutants are useful tools for studying a variety of biological processes, including flagellar motility, photosynthesis, and protein synthesis. Since Chlamydomonasspecies are normally haploid, the effects of mutations are seen immediately without further crosses.
In 2007, the complete nuclear genome sequence of C. reinhardtii was published.[4]
Channelrhodopsin-1 and Channelrhodopsin-2, proteins that function as light-gated cation channels, were originally isolated from C. reinhardtii.These proteins and others like them are increasingly widely used in the field of optogenetics.
Reproduction
Vegetative cells of the reinhardtii species are haploid with 17 small chromosomes. Under nitrogen starvation, vegetative cells differentiate into haploid gametes.There are two mating types, identical in appearance and known as mt(+) and mt(-), which can fuse to form a diploid zygote. The zygote is not flagellated, and it serves as a dormant form of the species in the soil. In the light, the zygote undergoes meiosis and releases four flagellated haploid cells that resume the vegetative lifecycle.
Under ideal growth conditions, cells may sometimes undergo two or three rounds of mitosis before the daughter cells are released from the old cell wall into the medium. Thus, a single growth step may result in 4 or 8 daughter cells per mother cell.
The cell cycle of this unicellular green algae can be synchronized by alternating periods of light and dark. The growth phase is dependent on light, whereas, after a point designated as the transition or commitment point, processes are light-independent.
Genetics
The attractiveness of the alga as a model organism has recently increased with the release of several genomic resources to the public domain. The Chlre3 draft of the Chlamydomonas nuclear genome sequence prepared by Joint Genome Institute of the U.S. Dept of Energy comprises 1557 scaffolds totaling 120 Mb. Roughly half of the genome is contained in 24 scaffolds all at least 1.6 Mb in length. The current assembly of the nuclear genome is available online.
The ~15.8 Kb mitochondrial genome (database accession: NC_ 001638) is available online at the NCBI database. The complete >200 Kb chloroplast genome is available online.
In addition to genomic sequence data, there is a large supply of expression sequence data available as cDNA libraries and expressed sequence tags (ESTs). Seven cDNA libraries are available online. A BAC library can be purchased from the Clemson University Genomics Institute. There are also two databases of >50 000 and >160 000 ESTs available online.
Evolution
Chlamydomonas has been used to study different aspects of evolutionary biology and ecology. It is an organism of choice for many selection experiments because (1) it has a short generation time, (2) it is both a heterotroph and a facultative autotroph, (3) it can reproduce both sexually and asexually, and (4) there is a wealth of genetic information already available.
Some examples (nonexhaustive) of evolutionary work done with Chlamydomonas include the evolution of sexual reproduction, the fitness effect of mutations,and the effect of adaptation to different levels of CO2.
According to one frequently cited theoretical hypothesis, sexual reproduction (in contrast to asexual reproduction) is adaptively maintained in benign environments because it reduces mutational load by combining deleterious mutations from different lines of descent and increases mean fitness. However, in a long-term experimental study of C. reinhardtii, evidence was obtained that contradicted this hypothesis. In sexual populations, mutation clearance was not found to occur and fitness was not found to increase.
DNA transformation techniques
Gene transformation occurs mainly by homologous recombination in the chloroplast and heterologous recombination in the nucleus. The C. reinhardtii chloroplast genome can be transformed using microprojectile particle bombardment or glass bead agitation, however this last method is far less efficient. The nuclear genome has been transformed with both glass bead agitation and electroporation. The biolistic procedure appears to be the most efficient way of introducing DNA into the chloroplast genome. This is probably because the chloroplast occupies over half of the volume of the cell providing the microprojectile with a large target. Electroporation has been shown to be the most efficient way of introducing DNA into the nuclear genome with maximum transformation frequencies two orders of magnitude higher than obtained using glass bead method.
Production of biopharmaceuticals
Genetically engineered Chlamydomonas reinhardtii has been used to produce a mammalian serum amyloid protein, a human antibody protein, human Vascular endothelial growth factor, a potential therapeutic Human Papilloma virus 16 vaccine,a potential malaria vaccine, and a complex designer drug that could be used to treat cancer.
Clean source of hydrogen production
Main article: Biological hydrogen production (Algae)
In 1939, the German researcher Hans Gaffron (1902–1979), who was at that time attached to the University of Chicago, discovered the hydrogen metabolism of unicellular green algae. Chlamydomonas reinhardtii and some other green algae can, under specified circumstances, stop producing oxygen and convert instead to the production of hydrogen. This reaction by hydrogenase, an enzyme active only in the absence of oxygen, is short-lived. Over the next thirty years, Gaffron and his team worked out the basic mechanics of this photosynthetic hydrogen production by algae.
To increase the production of hydrogen, several tracks are being followed by the researchers.
- The first track is decoupling hydrogenase from photosynthesis. This way, oxygen accumulation can no longer inhibit the production of hydrogen. And, if one goes one step further by changing the structure of the enzyme hydrogenase, it becomes possible to render hydrogenase insensitive to oxygen. This makes a continuous production of hydrogen possible. In this case, the flux of electrons needed for this production no longer comes from the production of sugars but is drawn from the breakdown of its own stock of starch.
- A second track is to interrupt temporarily, through genetic manipulation of hydrogenase, the photosynthesis process. This inhibits oxygen's reaching a level where it is able to stop the production of hydrogen.
- The third track, mainly investigated by researchers in the 1950s, is chemical or mechanical methods of removal of O2 produced by the photosynthetic activity of the algal cells. These have included the addition of O2 scavengers, the use of added reductants, and purging the cultures with inert gases. However, these methods are not inherently scalable, and may not be applicable to applied systems. New research has appeared on the subject of removing oxygen from algae cultures, and may eliminate scaling problems.
- A fourth track has been investigated, namely using copper salts to decouple hydrogenase action from oxygen production.[28]

No comments:
Post a Comment